Boolean Anatomy Search
Use the Boolean Anatomy Search application to perform complex queries for level of gene expression based on anatomical regions. There are two ways to perform boolean search of scored specimen expression: using the graphical user interface (GUI) or entering a text search string.
Table of Contents
- Finding the Boolean Anatomy Search
- Getting to know the Boolean Anatomy Search app
- Using the graphical user interface (GUI)
- Entering a search string
- Troubleshooting
Finding the Boolean Anatomy Search
From the top-level of the ATLAS-D2K website, click Data > Anatomy > Anatomy: Boolean Search.
You can also bookmark this link: https://www.atlas-d2k.org/deriva-webapps/boolean-search/
Getting to know the Boolean Anatomy Search app
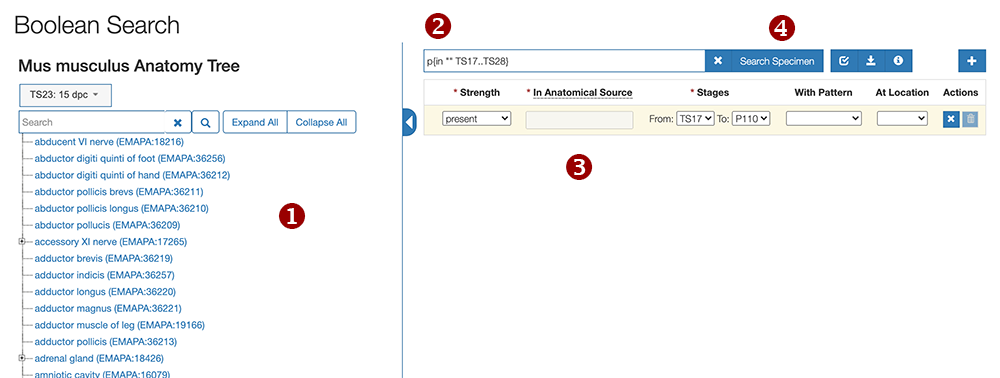
 |
Tree view of anatomy terms that conform to the GUDMAP Ontology. The dropdown button allows you to specify a Theiler Age Stage for the tree. The default is TS23 (15dpc). |
 |
Text field that displays the query as a search string. |
 |
Fields to add additional conditions to the query. |
 |
The Search Specimen button opens a search results page of all GUDMAP Specimen data that conforms to the query. |
Using the graphical user interface (GUI)
The application can visually represent queries as rows of search filters for each condition.
To build a query:
- Search and select an anatomical term for your first condition from the tree view in left panel. This will fill in the Anatomical Source field. Note: If you see no results for an anatomical region, make sure you are using the appropriate developmental stage in the dropdown box at the top of the anatomy tree.
- Select appropriate values for the dropdown fields:
- Strength: Choose from Not detected, Present, or Uncertain (required).
- Stages: Indicate a range of developmental stages (using Theiler Stages) (required).
- With Pattern: Choose from patterns of expression such as graded, homogenous, regional, etc.
- At location: Choose from various locations such as dorsal, lateral, etc.
- To add more conditions, click the plus (+) symbol on the top right corner.
- When you have built your query, click on the Search Specimen button. If all the values are valid, the resulting Specimen search results page will open in a new tab. If the query contains any invalid values, they will be highlighted in the table.
- Use the Save button to store the query in a text file. You may copy the contents and paste them into the text field to perform the query again.
Example of building a query with the GUI
Here is an example of building a query to find out which genes are expressed in the renal medullary interstitium, outer medullary interstitium and inner medullary interstitium in the adult mouse.
-
In the left hand section, above the search field is a dropdown list of different age stages. Click and select
TS28: Range P4-Adult. -
In the search field underneath, “renal medullary interstitium”. When you see the term appear in the tree, click the term to populate the condition box on the right side. Make sure the “Strength” field is set to
present.This is your first condition.
-
Click the plus (+) sign to add another condition and search for the term “outer medullary interstitium” and click to add your second condition.
-
Click the plus (+) sign one more time to add a third condition. Search for “inner medullary interstitium” and select it.
You should now have three condition on the right side of the screen:
-
Now click the “Search Specimen” button to search all gene records that show expression present in those regions:
-
In the search results, you’ll see in the Genes facet in the left sidebar the following genes: Crym, Wnt4, Ptch1. Select those facets and the search results will display specimen records with expression data related to those genes.
Here is a video demonstrating the above steps (note this demonstration was performed on the old GUDMAP website, but the functionality is the same on ATLAS-D2K):
Entering a search string
Alternatively, you can enter the entire query as a search string in the text field. You can then save this as a text file to download and save locally. Then at anytime you can copy the contents and paste them into the right-side text field to search again:
- Enter a query string consisting of one or more filters using the correct search syntax (see Search syntax below).
- Click the Validate button to check the validity of all values before submitting the query. This would also populate the table with any filters that were added directly in the text field.
- When you have built your query, click on the Search Specimen button. If all the values are valid, the resulting Specimen search results page will open in a new tab. If the query contains any invalid values, they will be highlighted in the table.
- Use the Save button to store the query in a text file. You may copy the contents and paste them into the text field to perform the query again.
Search syntax
Each filter should conform to the following syntax:
<strength>{in <anatomical source> <from stage>..<to stage> pt=<pattern> lc=<location>}
Multiple filters are combined using the keyword AND:
<strength>is one of the following symbols:pfor present,ndfor not detected, andufor uncertain.<anatomical source>is the anatomical name (e.g.gonad) shown in the tree view.<from stage>and<to stage>identify the search range of specimens. Specimens without stage information will not be included.<pattern>should be one of the symbols shown in the drop-down menu under Pattern in the GUI.<location>should be one of the symbols shown in the drop-down menu under Location in the GUI.
Example search strings
- One filter - where expression is detected in the bladder from TS17 through TS28 in the dorsal region:
p{in "bladder" TS17..TS28 pt=regional lc=dorsal} - Multiple filters - where expression is present in the artery and not detected in the arteriole from TS17 through TS28:
p{in "artery" TS17..TS28} AND nd{in "arteriole" TS17..TS28}
Troubleshooting
The following errors could occur when using the app:
- Incorrect formatting of the query in the text field. Please refer to the examples above.
- The Anatomical Source entered in the text field does not exist. This can be resolved by searching for the term in the tree view in the left pane, and use the exact term in the text field.
- Invalid values of Strength, Developmental Stages, Pattern or Pattern location. These can be resolved by selecting the proper values from the respective dropdown fields for each row.