Querying the ATLAS-D2K Data Browser
This section describes many ways to search ATLAS-D2K data:
Table of Contents
What is a data record in ATLAS-D2K?
A data record in ATLAS-D2K represents the metadata and links to the data files and other related records. Instead of just providing a title, abstract and files, ATLAS-D2K data records are organized into hierarchies based on the concepts used in research studies: Specimen, Study, Experiments and Replicates.
Every data record throughout ATLAS-D2K data has its own Resource IDentifier (RID). This is a permanent, citable, globally unique identifier displayed for every data record, much like an Accession Number.
The main top-level points of entry for most data in ATLAS-D2K are through the Specimen and Study records, described below.
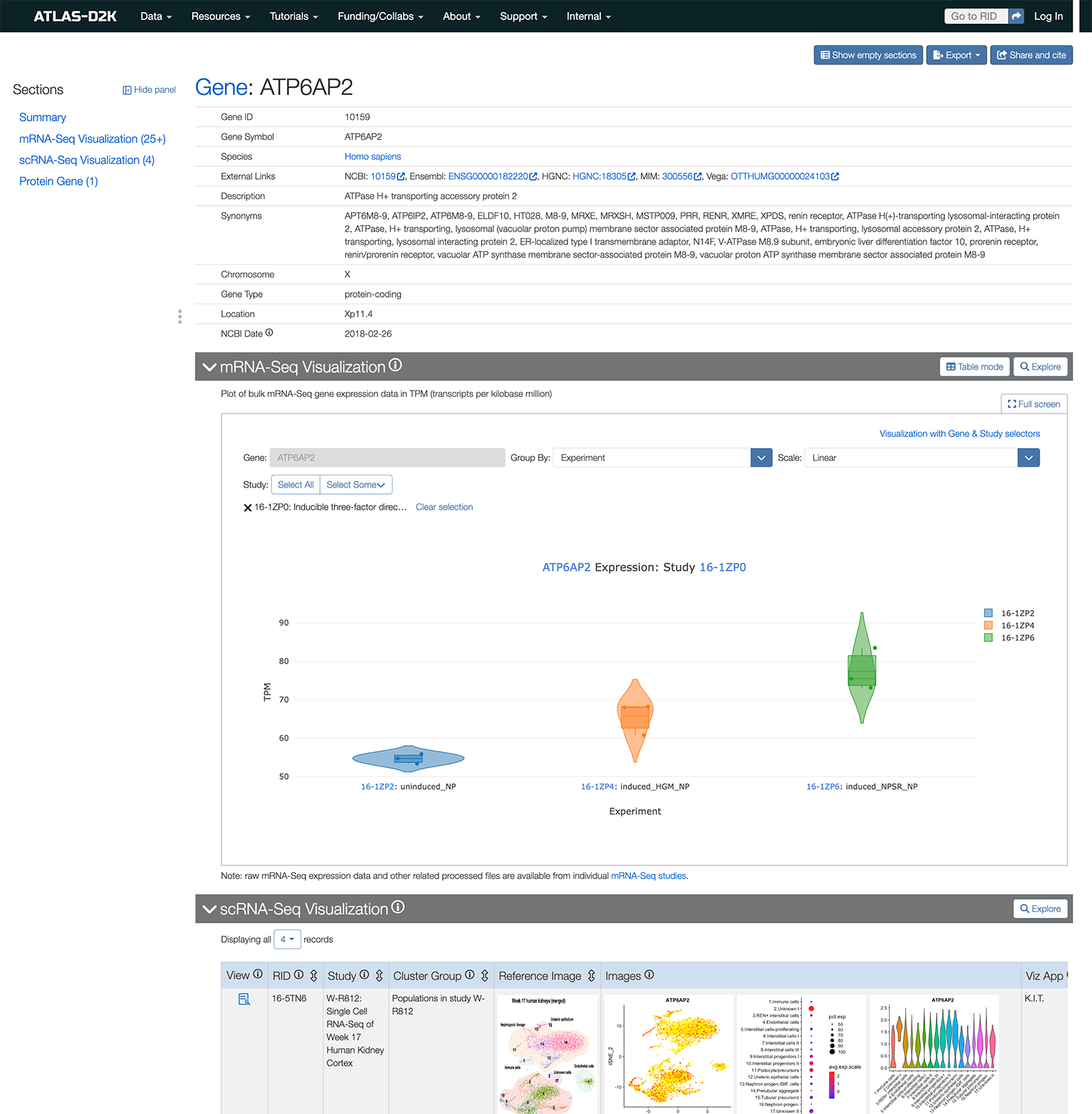
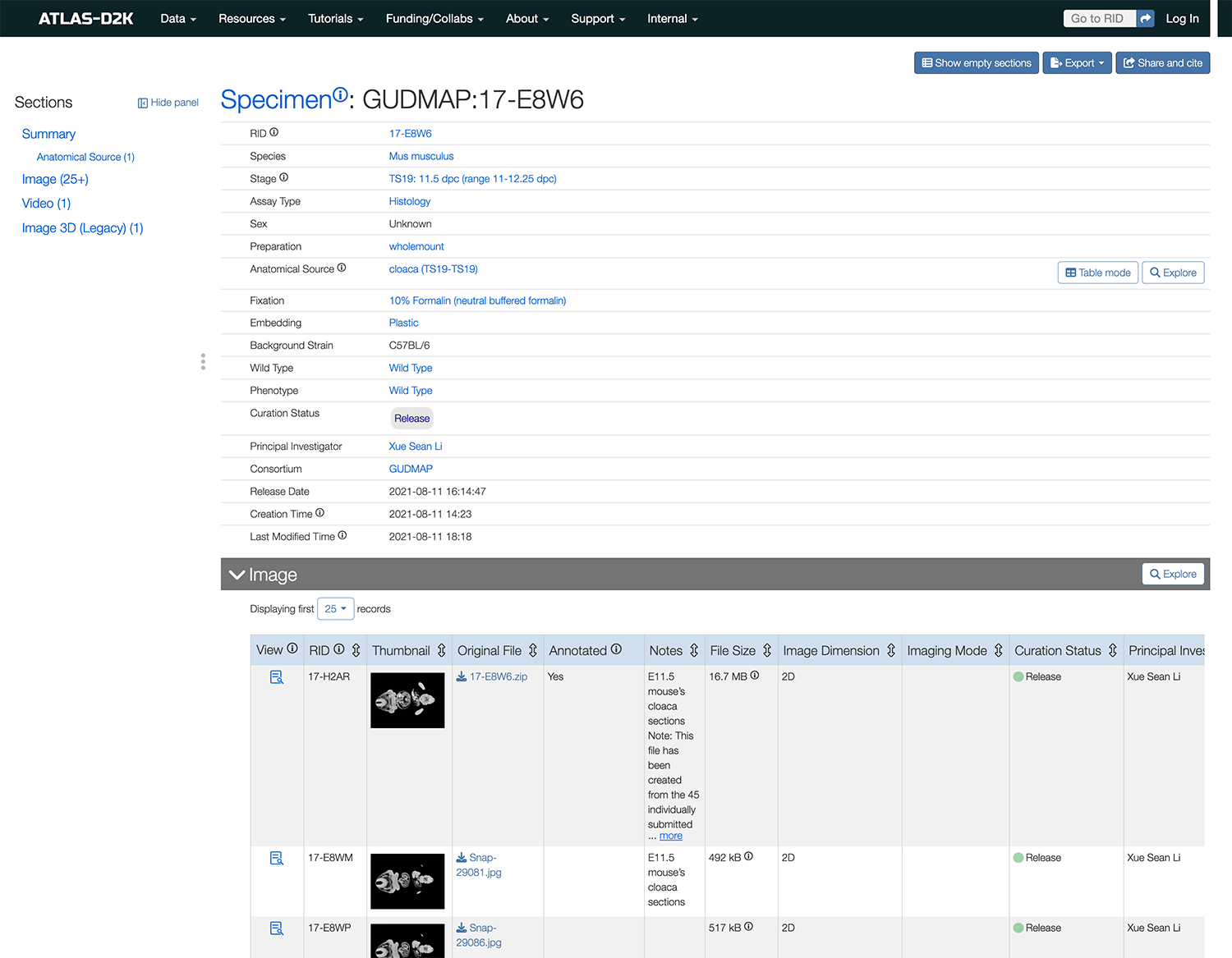
Specimen records (Imaging data)
A Specimen record describes the biological tissue from an organism (e.g., human, mice, organoid) used in experiments. In ATLAS-D2K, these records host imaging data and are linked to related sequencing studies (through Replicate records).
Specimen records also house information such as antibodies that were used, expression scores and probes (among others).
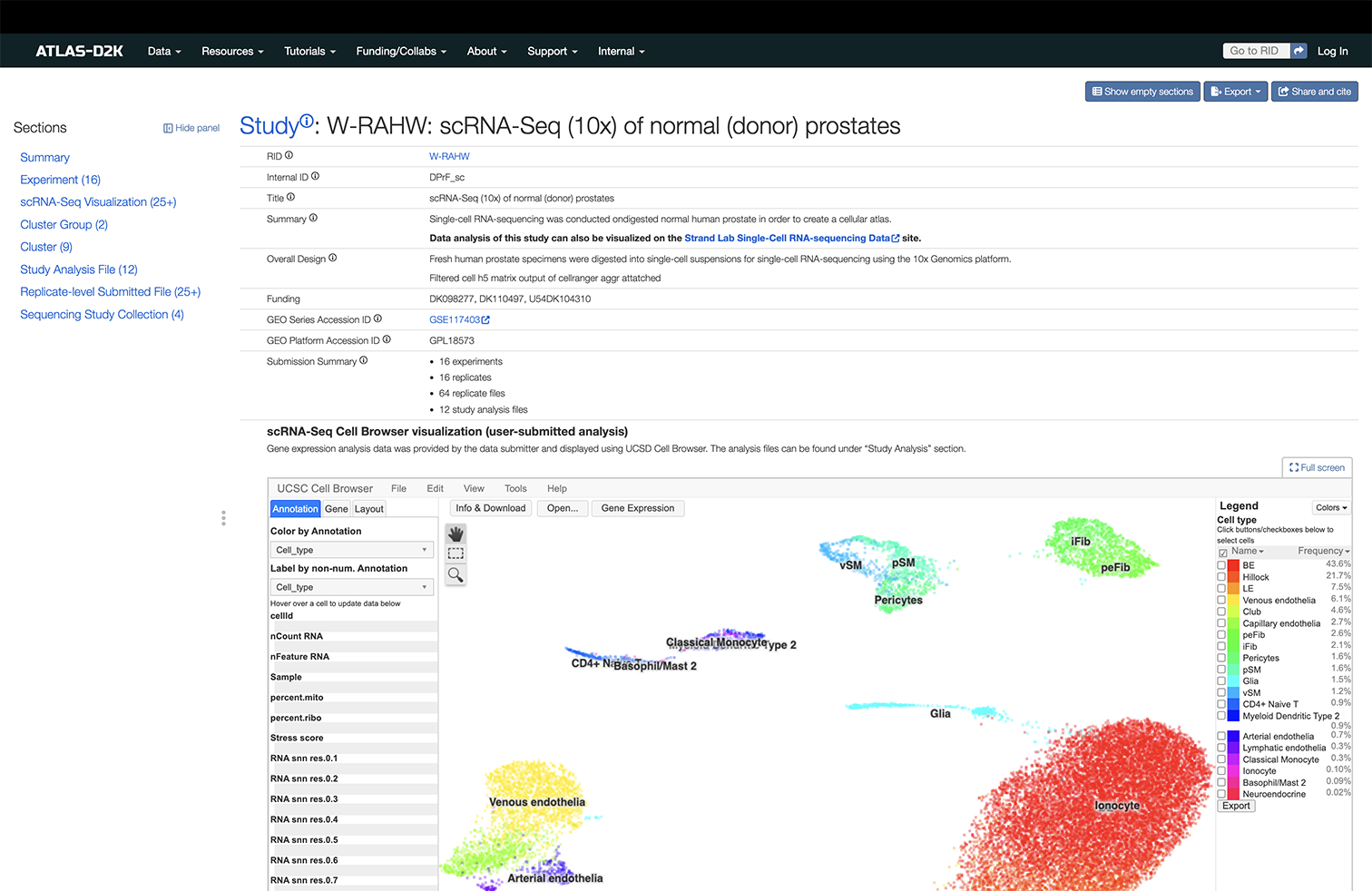
Study records (Sequencing/Omics data)
A Study is a group of Experiments, each of which consists of one or more Replicates, conducted for a specific scientific goal. Sequencing data is organized by Studies. The experiment types include -omic (e.g. transcriptomics, epigenomics, metabolomics) and Imaging Mass Cytometry (IMC). The data files can be found under individual Replicates or Studies. Legacy transcriptomics data can be found at Legacy Microarray and Legacy RNASeq.
Study records are organized as below:
- Study: The top layer, describes high-level objectives and overall design of the experiments. It may contain one or several Experiments. Study-level analysis files can be uploaded to the Analysis Files section.
- Experiment: Describes the protocols, procedures, and experiment settings done to a set of Replicates. Linked records could include antibodies, custom metadata, and settings.
- Replicate: Provides bio-sample details and both biological and technical replicate numbers. Replicates include:
- All of the replicate-specific experimental assays, such as sequencing and analysis files, are uploaded under the File section.
- A link to a Specimen record.
- For single cell RNA-Seq, you may also add a Single Cell Metrics record summarizing the statistics of a Replicate.
- Replicate: Provides bio-sample details and both biological and technical replicate numbers. Replicates include:
- Experiment: Describes the protocols, procedures, and experiment settings done to a set of Replicates. Linked records could include antibodies, custom metadata, and settings.
Query by Gene
Enter a gene symbol/name (or synonym) in the search box on the homepage (https://www.atlas-d2k.org) or by using the top-level menu navigation (Data > Gene).
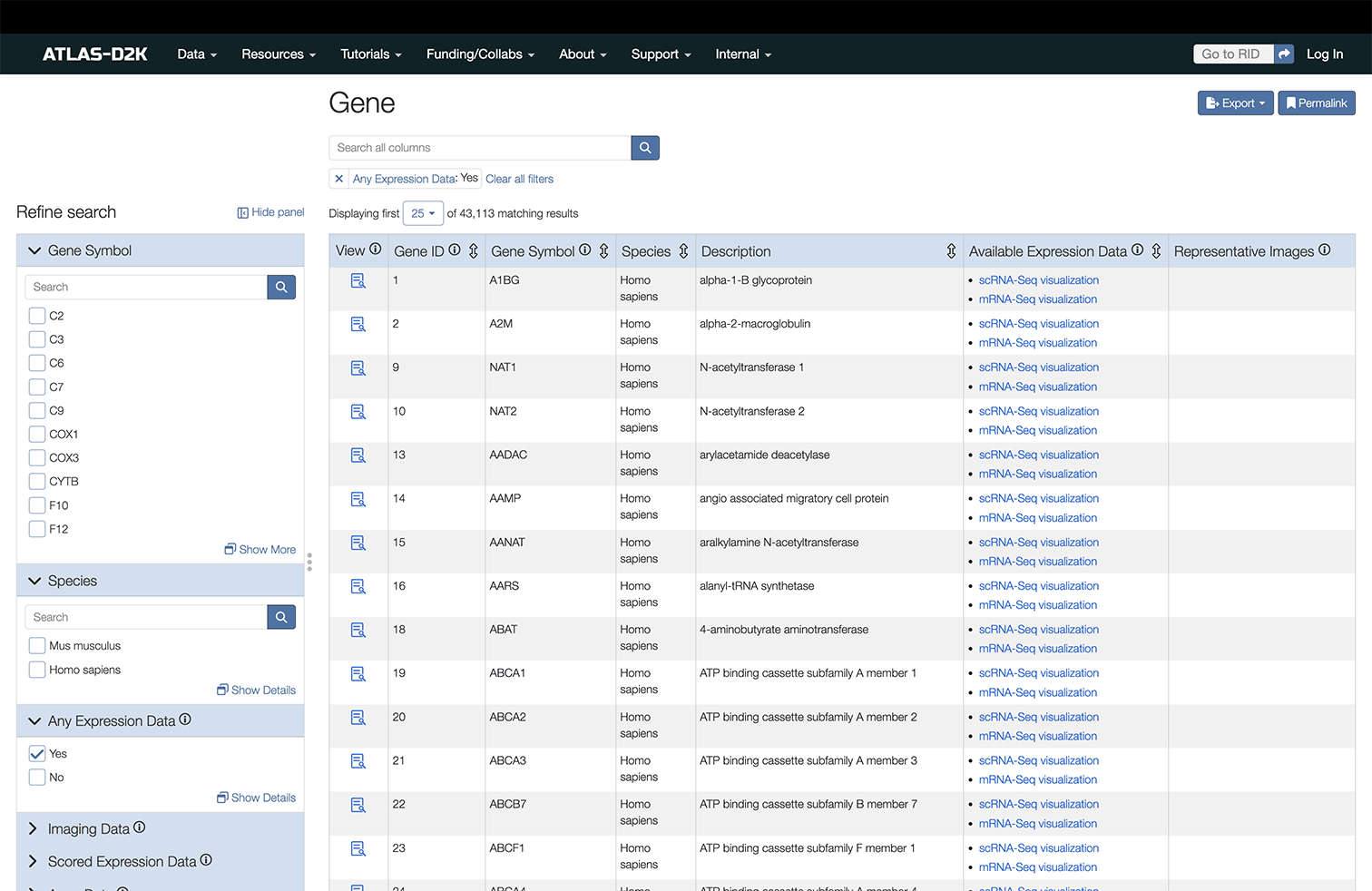
The results include records that contain information about the expression of the gene or genes of interest. The column “Available Expression Data” indicates the presence of Expression Scoring, Array Data or Imaging data from specimen (in situ, etc). The Imaging column includes representative thumbnails of the imaging data.
In the left sidebar of the results, you may filter results further by these categories:
-
Gene Symbol
-
Species
-
Synonyms
-
Any Expression Data (yes, no)
-
Imaging Data (yes, no)
-
Scored Expression Data (yes, no)
-
Array Data (yes, no)
-
scRNA-Seq Visualization (yes, no)
-
mRNA-Seq Visualization (yes, no)
-
Synonyms
-
MGI Symbol
-
Scored Anatomical Region
-
Specimen Assay Type
-
Antibody Tests
The search automatically assumes a ‘wildcard’ at the end of the search string; therefore, typing in ‘uro’ will search for words/symbols beginning with ‘uro’, e.g. urothelium, urogenital, uroplakin, etc.
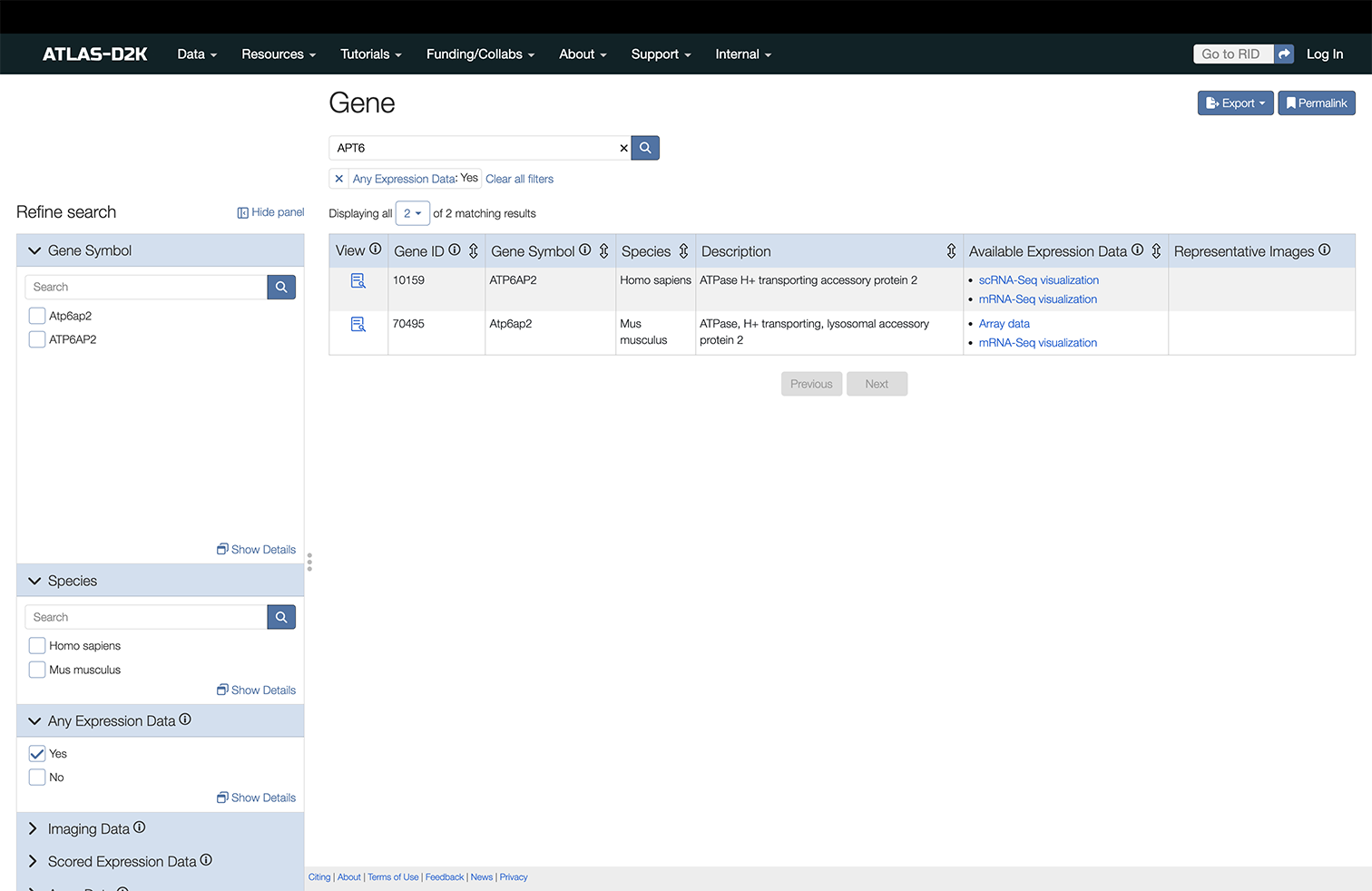
Multiple genes (Batch Query)
To search for multiple genes, use the Batch Query method. Use the search method described above but enter multiple genes separated with a pipe character ( | ) with no spaces in between. For example:
APT6|COX1|Six2
This list may contain a mixture of different terms (e.g. MGI accession IDs and MCBI Gene Symbols).
Querying with the Filtering Sidebar
This method guides your search through structured data by their classification or characteristics.
Biomedical research involves experiments on a group of model organisms (mice, zebrafish, etc) with particular age stages, there’s at least one anatomical region of interest, and there are different assays (RNA-Seq, microCT, etc). These are examples of the different categories or “facets” of the data.
When you perform a search in ATLAS-D2K, you’ll see the faceting navigation sidebar on the left, with the main search results on the right. As you explore the available facets and make choices, the main search results update to show what’s available with those combination of facets.
For example, if you are interested in sequencing data, you can go to the Study link in the top-level menu (Data > Study) and see a page that looks like this:
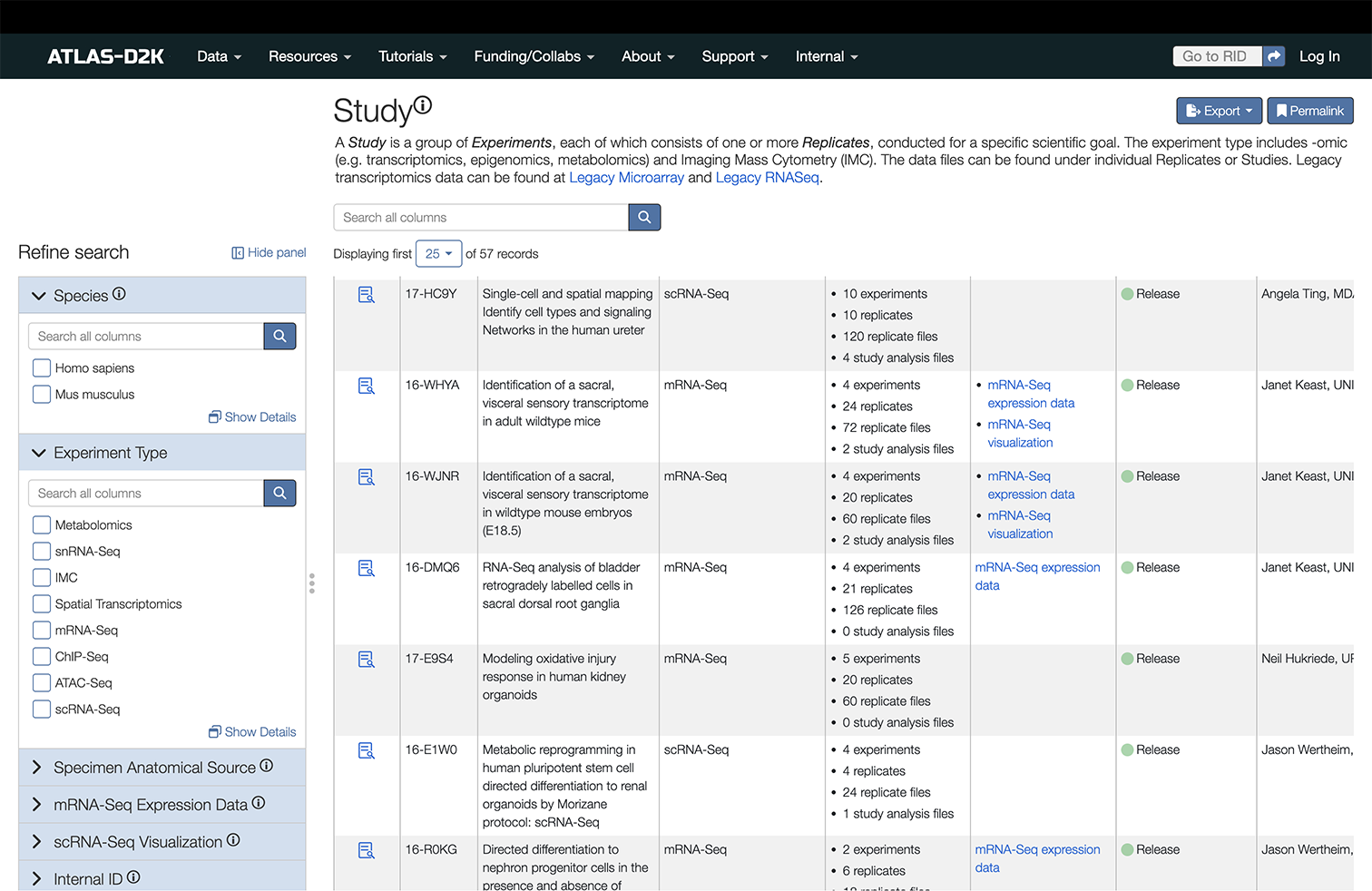
You can do a freetext search in the field above the results. And you can use the filtering sidebar in the left to narrow down the results even further.
Query by Anatomy
From the menu navigation, go to Data > Anatomy and choose either Anatomy: Facet Search or Anatomy: Tree View.
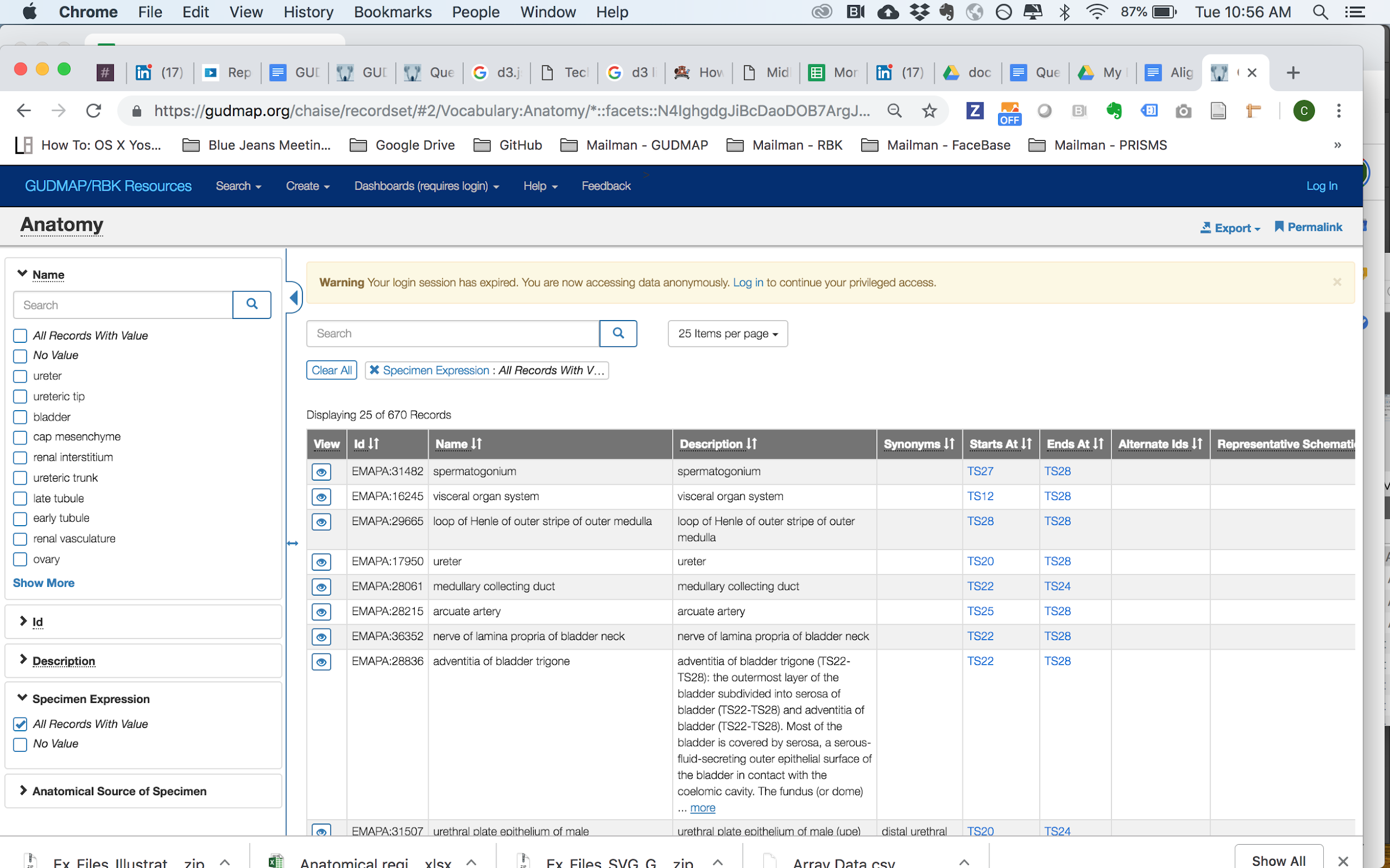
Searching by Anatomy Tree (for mouse anatomy)
Go to the Anatomy Tree view.
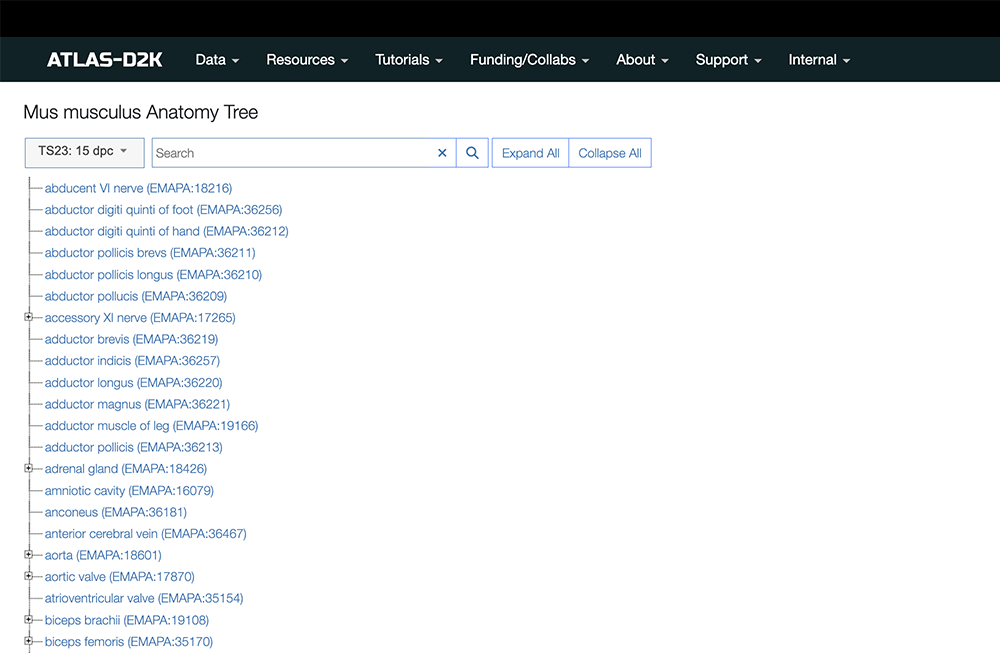
This option provides a tree structure view of anatomical terms that conform to the GUDMAP Ontology.
Start by choosing an Age Stage in the dropdown field and then typing an anatomical region in the search field.
In the example below, we chose “All Stages” and typed “prostate”. If there are results, the tree will highlight corresponding term(s) and scroll down to the first instance:
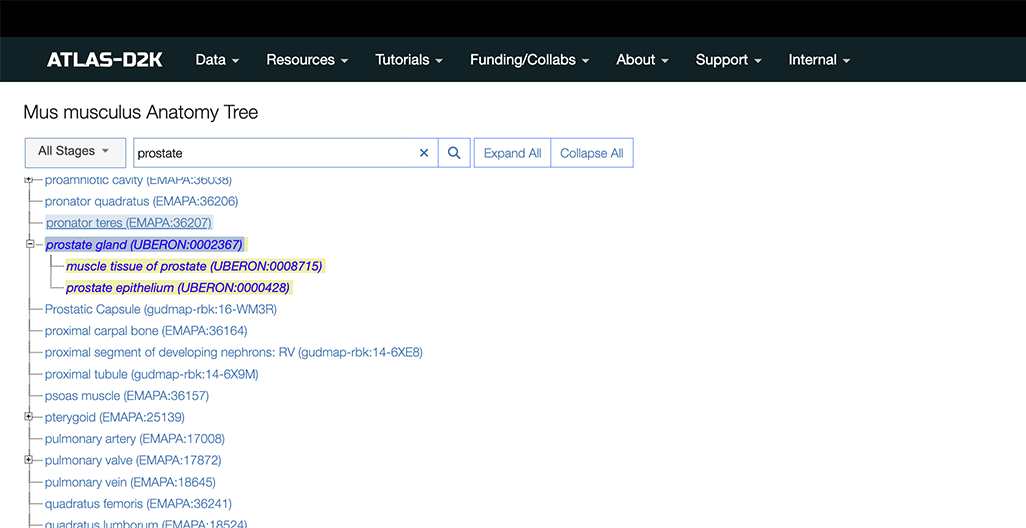
Click a term to go to the corresponding Anatomy record page (we have more details on this page in the next section):
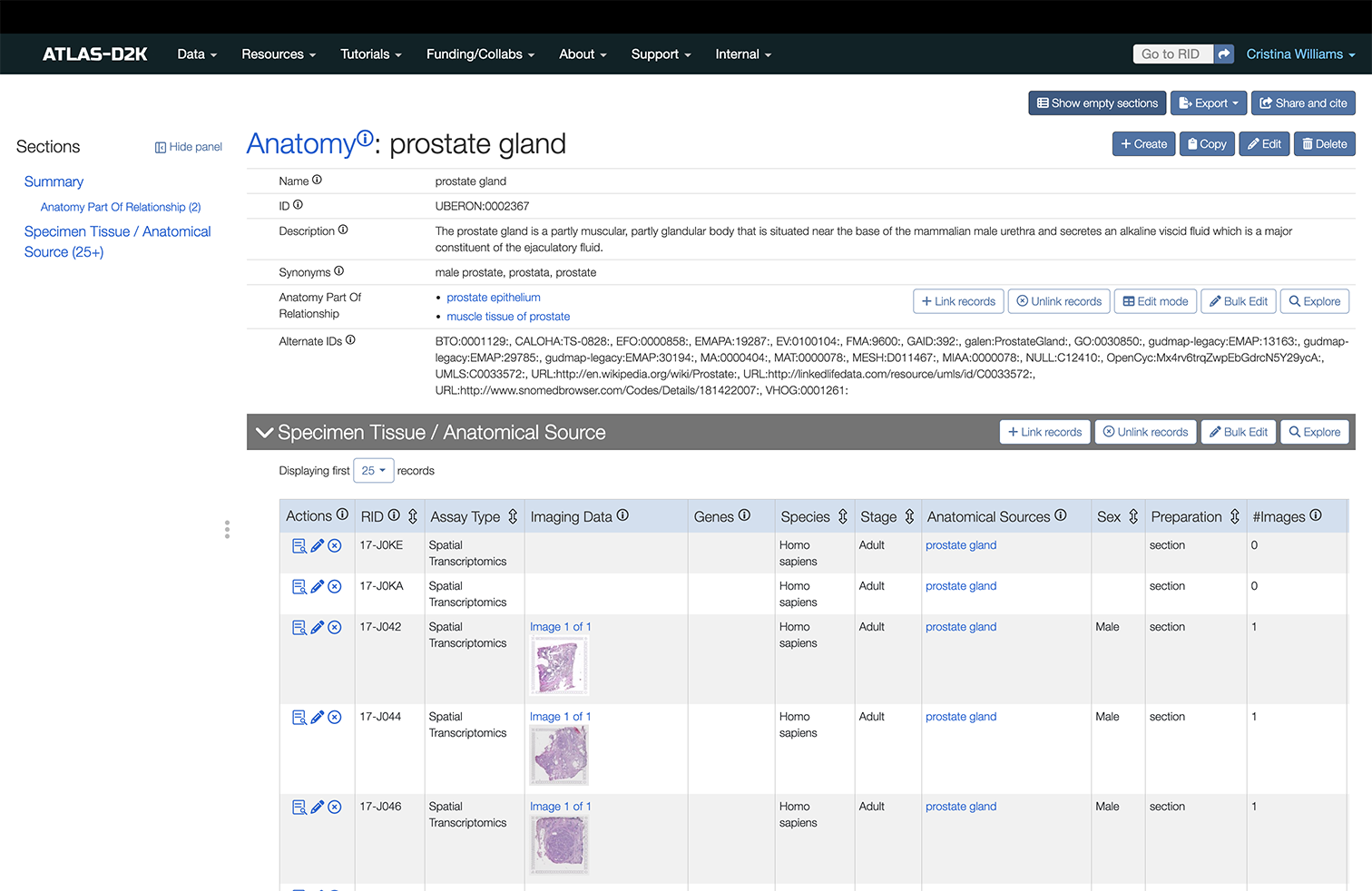
Anatomy: Faceted Search
You can reach the Anatomy faceted search page here.
This is the Anatomy section of the Data Browser where you may use the left filter sidebar to choose or search for anatomy regions by Name or ontology IDs (ie, EMAPA:18976). Click the View icon of the row you’re interested in to view the corresponding Anatomy record page (see below).
Anatomy record page
Below is an example of an Anatomy record page for the term “cortical renal tubule”.
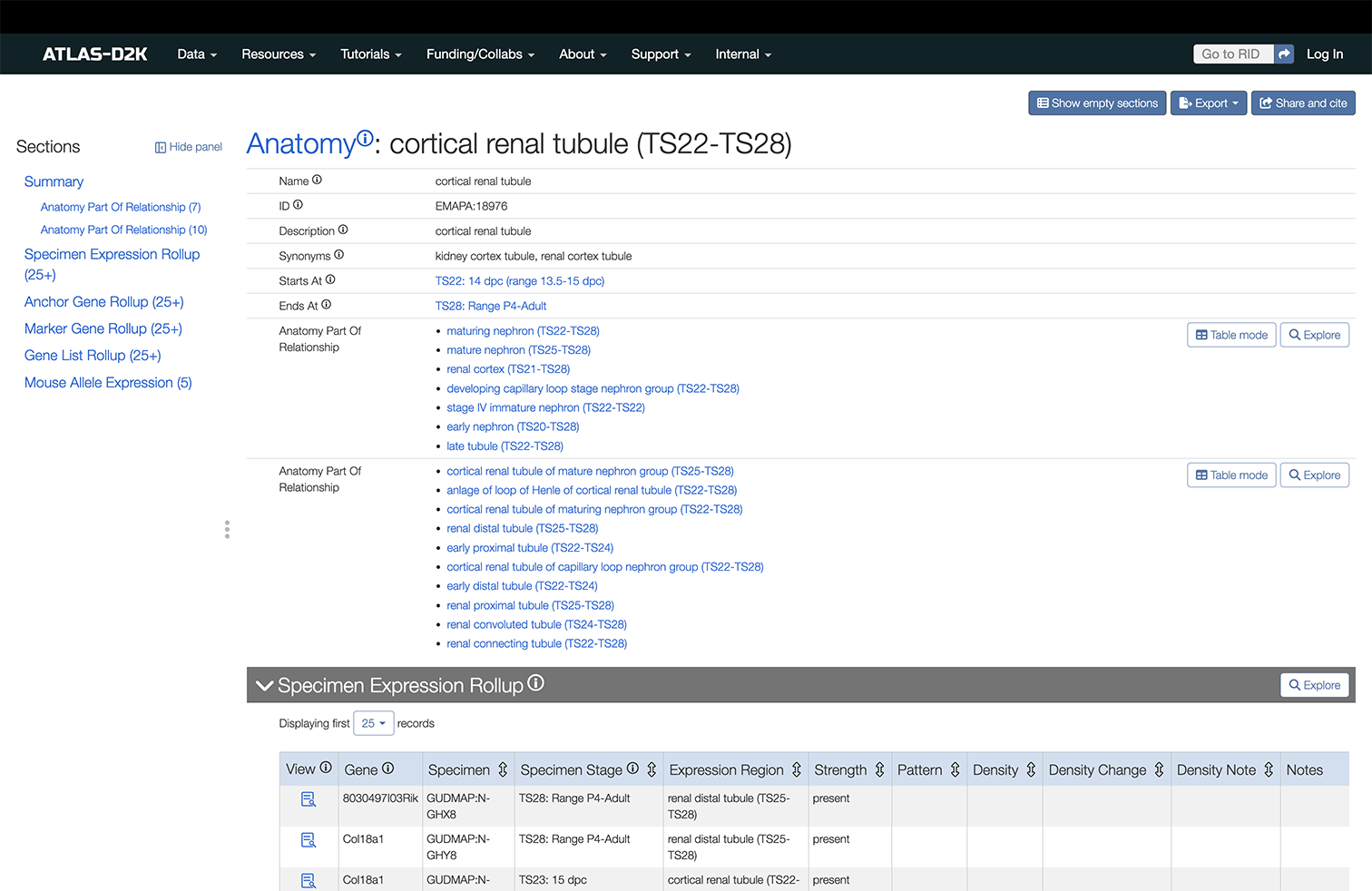
On the Sections sidebar to the right side of the page, you’ll see a list of available data including the following types:
-
Specimen Expression Rollup
-
Anchor Gene Rollup
-
Marker Gene Rollup
-
Gene List Rollup
-
Mouse Allele Expression
The numbers to the right of the names indicate how many instances of these types of data are available.
Search for genes annotated with expression present, uncertain or not detected
You can search sequencing data for annotations on expression strength.
From an Anatomy record page, choose “Specimen Expression Rollup” in the Sections list. This table includes entries that include annotated in situ expression data and microarray data.
-
For Specimen (ISH/IHC) that contain either a direct annotation for the anatomical term specified (whether it is present, uncertain or possible) or an inferred annotation for the anatomical term (see below).
Each entry includes the relevant symbol for the gene expressed in the queried structure. Symbols are the current standard gene symbols (see NCBI). To better understand the context of your results please see the tutorial on genitourinary development.
-
For Microarray where the anatomical term specified is the sample material or where sub-components of the anatomical term have been used as sample material.
For example, using the term ‘maturing nephron’ for the query will return database entries where the sample was the ‘early proximal tubule’ and entries where the sample was ‘maturing renal corpuscle’. Both structures are part of the maturing nephron.
Challenges on searching by anatomical terms
Structures are often referred to with a variety of different terms. The predictive text in the “Tissue (Anatomical Source)” filter on the Specimen search page will help you enter the correct term.
However, some structures have common names that begin with different text from the ontology name. For example, proximal tubule is represented by the term renal proximal tubule in the ontology.
You can easily find the ontology term for a structure by viewing the interactive anatomy ontology tree on the left side of the Boolean Anatomy Search page. This tree is supported by a text string search that will find terms containing a given string. For example, typing proximal in the “find anatomy component” box will highlight renal proximal tubule in the tree.
Queries can be performed for multiple components by entering terms separated by a pipe | character with no space in between (e.g. kidney|ovary). Predictive text is available only for the first term in the list but other valid ontology terms can be added by typing on, using the pipe character to separate terms. Queries with multiple terms are treated as A OR B OR C.
Inferred annotation
We support inferred annotation, that is, we indicate if a region inherits the annotation of a related region.
Suppose, for example, the anatomical term “superficial cellular layer” has been annotated as expression present for a particular gene.
As a consequence, the anatomical term “urothelium” has the inferred present annotation (even though it has not been annotated directly) because “superficial cellular layer” is a part of the urothelium. Equally, if urothelium was annotated as not detected then its parts, including superficial cellular layer would have the inferred not detected annotation.
The original annotations and original expression images are displayed on the page for the corresponding database entry.
Boolean Anatomy Search
The Boolean Anatomy Search allows complex queries to be constructed to search for gene expression based on selected anatomical structures.
The search allows combinations of structures and developmental stages and different combinations of expression found to be present, not detected and uncertain. The search can be applied to a combination of structures or just to one structure.
For example, to retrieve only genes expressed in a structure, or to compare expression in the same structure at different stages.
For more details, please go to the Boolean Anatomy Search page.