Exporting Data
On this page:
- 1. Export Options
- 2. Export the BDBAG
- 3. Export the data files
- 4. Exporting to submit to GEO
- 5. Troubleshooting
1. Export Options
Available data files displayed on a page may be downloaded individually by clicking the link, but the “Export” button offers many more options:
- This record (CSV): Available on every page, this option downloads a CSV file of the metadata. For search results, it will include metadata for all of the results, not just those visible on the current page.
- BDBAG: Available only on detail pages. To download the data files, this must be used with our tool. This option downloads a ZIP file with metadata and information about the files you want to download. This ZIP file must be used with our tool, see below, to export the files to your desired location (locally or on a remote server). This is recommended for many files or very large files.
- GEO Excel: Available on Study detail pages only, this option exports an Excel spreadsheet with metadata suitable for uploading to GEO.
- GEO BDBAG (GEO excel and sequencing files): Available on Study detail pages only, this option exports an Excel spreadsheet with metadata and a BDBAG ZIP file, which must be used with our tool, see below, that is suitable for uploading to GEO.
1.1 What is a BDBAG?
BDBAG (Big Data Bag) is a program that allows for reliable transfer of a “bag” of digital content - in this context, a group of files that you want to export in bulk. It is available as a command-line client and a GUI client.
A BDBAG consists of a hierarchical directory containing the data files and other accompanying metadata files that enable and document the secure transfer of the BDBAG to your chosen location. This provides verification that you have all the files you were trying to export AND that they were not corrupted in the process.
The rest of this document describes how to use the BDBAG export with our tool to export data to your desired destination.
2. Export the BDBAG
To export the initial “BDBAG”, go to the detail page with the data you want, click the Export button towards the upper right part of the screen and choose BDBAG.
We’ll use this study as an example: https://www.atlas-d2k.org/chaise/record/#2/RNASeq:Study/RID=16-WW2M
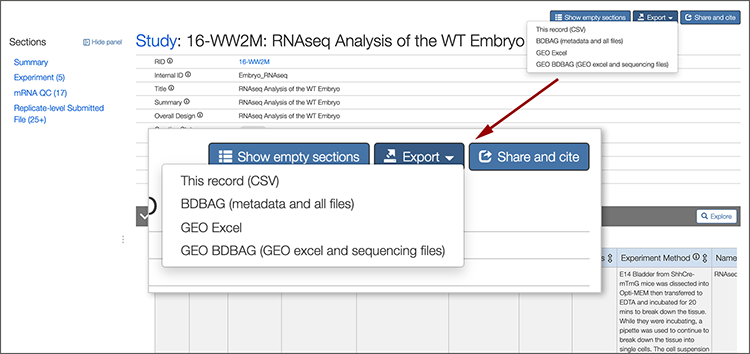
A progress window appears saying that the BDBAG is being created.
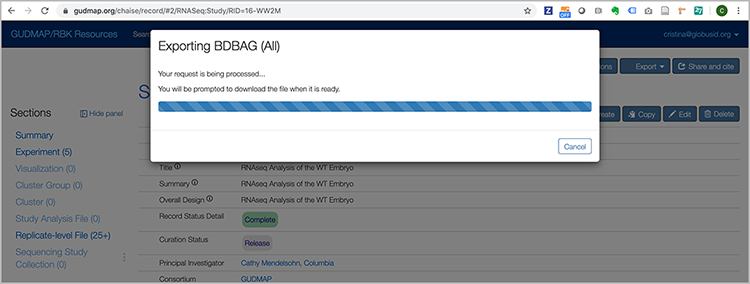
Then a ZIP file downloads to your local environment with the metadata files describing the data (including appropriate MD5 checksums), e.g. Study_16-WW2M.zip. This will not include the actual data files themselves.
Make a note of the path to this file - it will be used in the export tool to fetch the files.
3. Export the data files
Once you have downloaded and installed the client tools (see below), use our BDBag client to export the files to a destination of your choice.
You have the option of using a GUI for simple transfer of files to your local computer or the command-line client, which offers more options and may be used on a remote server.
3.1 Download and install DERIVA Client tool
DERIVA Client is a set of software packages that includes the BDBag command line program (bdbag) and a GUI, either of which may be used for downloading the files, and DERIVA Authentication Agent for authenticating your downloads. (Note that there are other programs within this package that are useful for data submitters, such as the upload program.)
- Windows and Mac users or those who prefer a more “turnkey” installation: installers are available here.
- Linux users: DERIVA Client is built on Python and is available via the command
pip install deriva-client. A Python 3.5.4 or greater system installation is required. The latest stable version of Python is recommended.
Detailed installation instructions are available here.
3.2 Exporting files using the GUI
The GUI application is quick and easy to use but has fewer options than the command-line. This is fine if you just want all of the files from the BDBag downloaded to a local environment. For use on a remote server or to have full access to exporting options, see 3.3 Exporting files using the command line client.
To use the GUI:
-
Go to the application folder where you installed the client tools (The directory name will be “DERIVA Client Tools”). Double-click the icon for BDBag to open it.
-
In the Name column, use the arrows to traverse down your system directory to the folder with the BDBag you downloaded (ie, your Downloads folder). Select the BDBag zip file. The following screenshots provide a demonstration. But keep in mind that if you traverse to a folder with many files, it will take a while to scroll to the BDBag file. You may want to save it in a more “shallow” location.
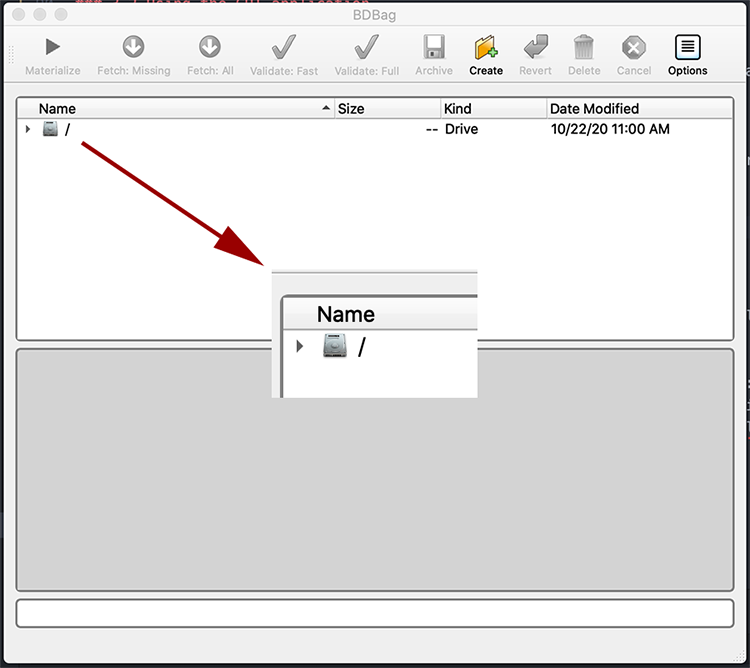
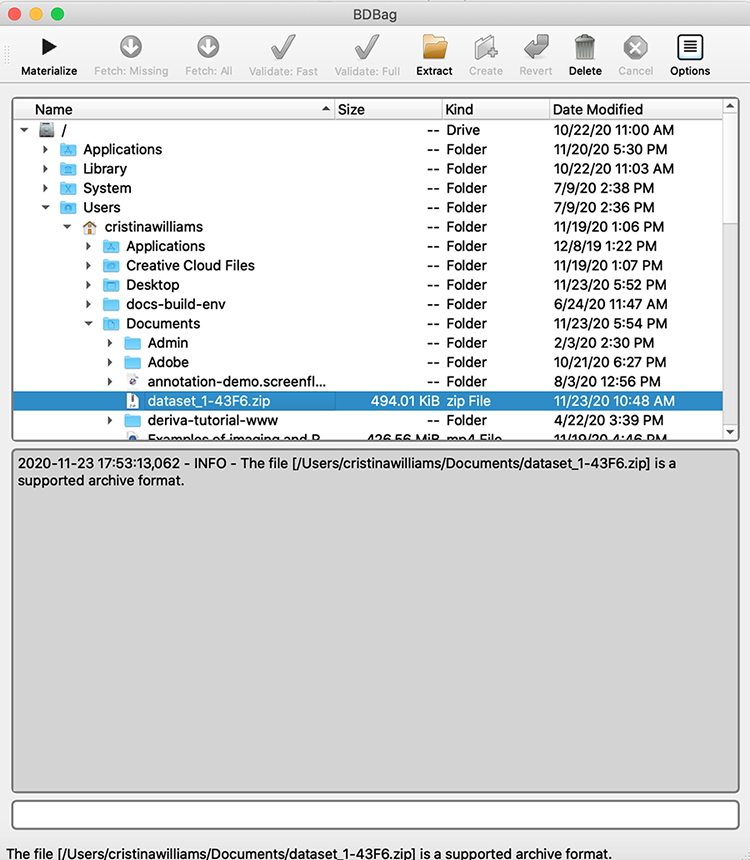
- Click Materialize to extract the folder from the zip file, download them to the same folder as the zip file and perform checksum validation to make sure there was no corruption in the transfer. The status field will indicate your progress.
3.3 Exporting files using the command line client
Using the command line client, bdbag, requires more steps but offers many more options. In general, the steps are:
- Authenticate with our Authentication Agent.
- Use the DERIVA Command Line Applications terminal window to run the appropriate
bdbagcommand, e.g.,bdbag download\path\Study_16-WW2M.zip --materialize
3.3.1 Log in with DERIVA Authentication
Open the DERIVA Authentication application:
- Windows or Mac: Go to the applications folder, open the “DERIVA Client Tools” folder and double-click the icon for “DERIVA Authentication Agent”.
- Linux: Run
deriva-auth.
The first time you log in, you’ll see a mostly-empty window:
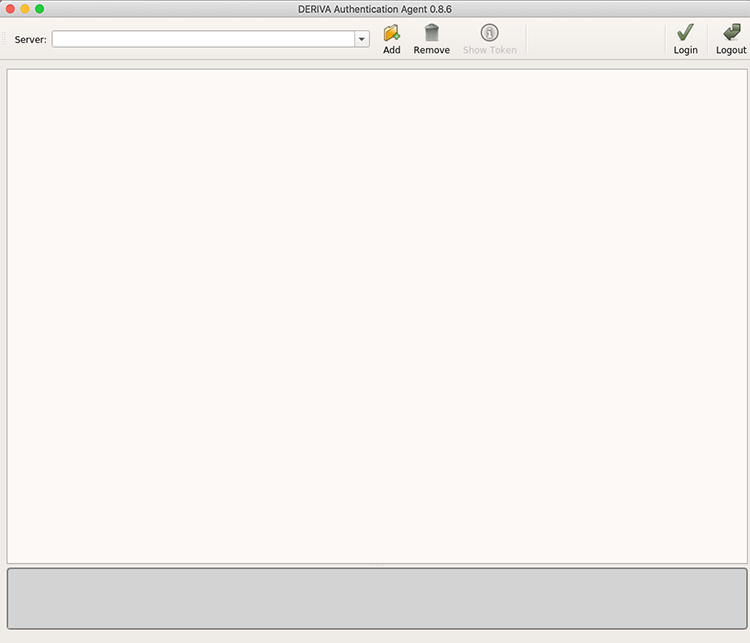
In the “Server:” field, type in www.atlas-d2k.org and click Enter or click the Add button.
You should now see the following screen:
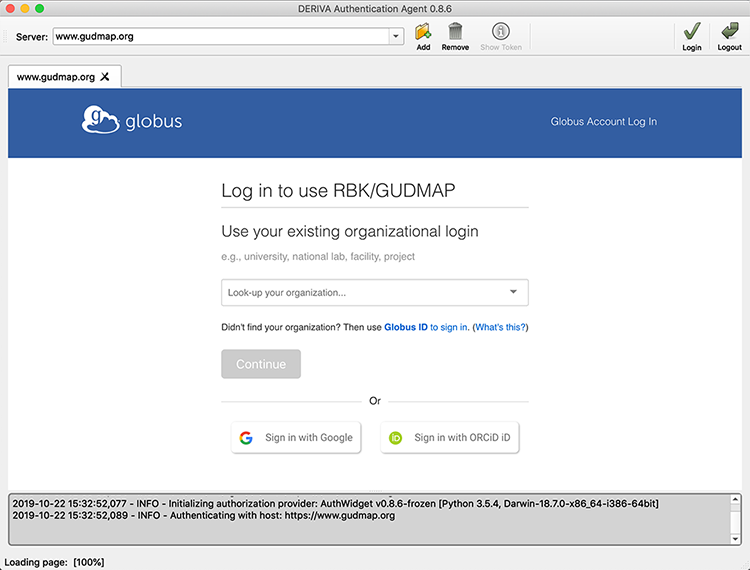
Use the log in method of your choice (i.e., your institution’s credentials if available).
After logging in, you’ll see an “Authentication Successful” message:
3.3.2 Download the data files
The following describe two methods of downloading the data files: to your local computer and to a remote server.
3.3.2.1 Download to your local computer
If you used a Window/Mac installer to install DERIVA Client, go to the “DERIVA Client Tools” folder on your computer and open “DERIVA Command Line Applications”. This will display a special Terminal window to run your BDBag commands.
Run the following command to export (i.e., materialize) the data and download them to the current directory:
bdbag download\path\Study_16-WW2M.zip --materialize
This program outputs messages as it extracts the bag, looks up the remote addresses of the data files and downloads each file. This program also automatically performs full validation of the files after they have been downloaded.
The downloaded files are located in one or more sub-folders under ./data such as ./data/asset/ or ./data/files/; all these folders are in the materialized directory.
3.3.2.2 Download to a remote cluster/server
If you want to download data files to a remote cluster or server, you will need to install the DERIVA Client tools on the remote environment as well as locally (as described in 3.1 Download and install DERIVA Client).
A Python 3.5.4 or greater system installation is required to use the client tools. The latest stable version of Python is recommended.
-
Install DERIVA Client on the remote cluster/server as well as locally. We recommend using the instructions for Installing
deriva-clientfrom PyPi viapip. - Get a token: On your local computer, run:
deriva-globus-auth-utils login --host <hostname> --no-local-serverwhere
<hostname>is the server hosting the data (ie,www.atlas-d2k.org). -
A browser window will open asking for your consent to authenticate using Globus. Click “Allow”.
-
Copy the displayed authorization code and go back to the
deriva-globus-auth-utilscommand in your Terminal window and paste in the code as instructed. You should see aLogin successfulmessage. -
Copy the BDBag zip file (e.g. “Study_16-WW2M.zip”) to the remote server.
- SSH to the remote server and run the following command to export (i.e., materialize) the data and download them to the current directory:
bdbag download\path\Study_16-WW2M.zip --materialize
3.3.3 More bdbag options
You may refine the download options further with the following options. To use these options, first unzip the downloaded file.
unzip download\path\Study_16-WW2M.zip
This command will unzip the file and create a directory in your current folder.
3.3.3.1 Download only files with a particular file extension
To download only the fastq files (.fastq.gz) from a study:
bdbag --resolve-fetch all --fetch-filter 'filename$*fastq.gz' download\path\Study_16-WW2M
3.3.3.2 Download only the missing files with a particular file extensions
Similar to the above command, but you would use the option --resolve-fetch missing if your download quit before retrieving all of the files.
bdbag --resolve-fetch missing --fetch-filter 'filename$*fastq.gz' download\path\Study_16-WW2M
3.3.3.3 Filter and download only files below a specific size
You can specify a file size limit for your download:
bdbag --resolve-fetch all --fetch-filter 'length<=n' download\path\Study_16-WW2M
where n is the size limit in kilobytes.
For example, if you only wanted to download files smaller than 500 MB, the command would be:
bdbag --resolve-fetch all --fetch-filter 'length<=500000' download\path\Study_16-WW2M
3.3.4 All bdbag command line options
You may find the complete list of command line options here:
https://github.com/fair-research/bdbag/blob/master/doc/cli.md#bdbag-command-line-interface-cli
4. Exporting to submit to GEO (for Data Submitters)
ATLAS-D2K members can upload their study to ATLAS-D2K and then export the study in a GEO-compliant package. This requires downloading and installing DERIVA Client to your local environment if you haven’t done so already.
- Create the study in ATLAS-D2K.
- From the Study page, click Export and choose
GEO BDBAG (GEO excel and sequencing files). This will create a zip folder that has an Excel spreadsheet containing metadata and file locations. ExportGEO Excelto check the excel content that will be included in theGEO BDBAG. - Use the
bdbagcommand to export all those files:bdbag download\path\Study_16-WW2M.zip --materializeAlternatively, if the files are local, simply unzip the GEO BDBAG zip file and copy your files into the folder.
- Zip the directory and send this file to GEO.
FTP files to GEO
GEO supports multiple ftp client software including lftp. You can use the graphical user interface tools to transfer the bag folder. This section provides simple instructions using lftp on linux which is a common OS for cluster environment.
After you create an account with GEO. They will provide you the following information for data submission:
- ftp server e.g. ftp-private.ncbi.nlm.nih.gov
- user name
- password
- upload directory related to your submission e.g.
uploads/barmfield@orcid_4vEVKoyE
To upload files to GEO:
- login to the ftp server
> lftp -u <user name> ftp-private.ncbi.nlm.nih.gov`This will prompt you for the password. Enter the password. After the password is entered correctly, it will give you a prompt to the ftp server.
- Run the following command to transfer files from your local folder to the ftp server
2.1 cd to your download upload directory
> cd uploads/barmfield@orcid_4vEVKoyE
2.2 mirror your local folder with the remote ftp folder. When run the mirror command, specify the path to the completed bag folder. The mirror will try to replicate what’s in the local folder to the remote site. See this site for more instructions and examples.
> mirror -R /path/to/your/bag/Study_17-DS7J Study_17-DS7J
You can also upload individual files using put or mput (for multiple files). Use mkdir to create directories at the remote site.
- Check that all the files are downloaded properly. Use
cdandlscommand to change directory and list files in the directories to make sure that all the files are there. e.g.> cd Study_17-DS7J > lsIf everything is there, then the submission is done. When notify the GEO submission, indicate that all relevant files are under the Study_17-DS7J/data folder.
- To list all available commands, type help.
> help
5. Troubleshooting
5.1 If you used DERIVA Auth but are still getting an error that you are not allowed to download data
Make sure that you logged in to the right server! This is a common mistake if you are using this procedure on different websites. Make sure to double-check the content of the “Server” field in DERIVA Auth.
5.2 If your BDBAG export failed
BDBag keeps track of what files were exported. If the transfer is interrupted, it will re-try several times.
If the transfer completely stops, you try again in the GUI or if you’re using the commandline, rerun the same command with the option --missing. Either way, the client tool will only get the appropriately missing files.
5.3 If you want to verify you received all of the files in the BDBAG
Re-run your BDBag command with --validate full at the end to confirm the data integrity.